The first sourmash tutorial - making signatures, comparing, and searching¶
You’ll need about 5 GB of free disk space, and about 5 GB of RAM to search GenBank. The tutorial should take about 20 minutes total to run. In fact, we have successfully tested it on binder.pangeo.io if you want to give it a try!
Install sourmash¶
You’ll need to install sourmash first!
Generate a signature for Illumina reads¶
Download some reads and a reference genome:
mkdir ~/data
cd ~/data
curl -L https://osf.io/ruanf/download -o ecoliMG1655.fa.gz
curl -L https://osf.io/q472x/download -o ecoli_ref-5m.fastq.gz
Compute a scaled signature from our reads:
mkdir ~/sourmash
cd ~/sourmash
sourmash sketch dna -p scaled=10000,k=31 ~/data/ecoli_ref*.fastq.gz -o ecoli-reads.sig
Compare reads to assemblies¶
Use case: how much of the read content is contained in the reference genome?
Build a signature for an E. coli genome:
sourmash sketch dna -p scaled=1000,k=31 ~/data/ecoliMG1655.fa.gz -o ecoli-genome.sig
and now evaluate containment, that is, what fraction of the read content is contained in the genome:
sourmash search ecoli-reads.sig ecoli-genome.sig --containment
and you should see:
select query k=31 automatically.
loaded query: /home/jovyan/data/ecoli_ref-5m... (k=31, DNA)
loaded 1 signatures.
1 matches:
similarity match
---------- -----
31.0% /home/jovyan/data/ecoliMG1655.fa.gz
Try the reverse, too!
sourmash search ecoli-genome.sig ecoli-reads.sig --containment
Make and search a database quickly.¶
Suppose that we have a collection of signatures (made with sourmash compute as above) and we want to search it with our newly assembled
genome (or the reads, even!). How would we do that?
Let’s grab a sample collection of 50 E. coli genomes and unpack it –
mkdir ecoli_many_sigs
cd ecoli_many_sigs
curl -O -L https://github.com/sourmash-bio/sourmash/raw/latest/data/eschericia-sigs.tar.gz
tar xzf eschericia-sigs.tar.gz
rm eschericia-sigs.tar.gz
cd ../
This will produce 50 files named ecoli-N.sig in the directory ecoli_many_sigs/ –
ls ecoli_many_sigs
Let’s turn this into an easily-searchable database with sourmash index –
sourmash index ecolidb ecoli_many_sigs/*.sig
and now we can search!
sourmash search ecoli-genome.sig ecolidb.sbt.zip -n 20
You should see output like this:
select query k=31 automatically.
loaded query: /home/ubuntu/data/ecoliMG1655.... (k=31, DNA)
loaded 0 signatures and 1 databases total.
49 matches; showing first 20:
similarity match
---------- -----
75.9% NZ_JMGW01000001.1 Escherichia coli 1-176-05_S4_C2 e117605...
73.0% NZ_JHRU01000001.1 Escherichia coli strain 100854 100854_1...
71.9% NZ_GG774190.1 Escherichia coli MS 196-1 Scfld2538, whole ...
70.5% NZ_JMGU01000001.1 Escherichia coli 2-011-08_S3_C2 e201108...
69.8% NZ_JH659569.1 Escherichia coli M919 supercont2.1, whole g...
59.9% NZ_JNLZ01000001.1 Escherichia coli 3-105-05_S1_C1 e310505...
58.3% NZ_JHDG01000001.1 Escherichia coli 1-176-05_S3_C1 e117605...
56.5% NZ_MIWF01000001.1 Escherichia coli strain AF7759-1 contig...
56.1% NZ_MOJK01000001.1 Escherichia coli strain 469 Cleandata-B...
56.1% NZ_MOGK01000001.1 Escherichia coli strain 676 BN4_676_1_(...
50.5% NZ_KE700241.1 Escherichia coli HVH 147 (4-5893887) acYxy-...
50.3% NZ_APWY01000001.1 Escherichia coli 178200 gec178200.conti...
48.8% NZ_LVOV01000001.1 Escherichia coli strain swine72 swine72...
48.8% NZ_MIWP01000001.1 Escherichia coli strain K6412 contig_00...
48.7% NZ_AIGC01000068.1 Escherichia coli DEC7C gecDEC7C.contig....
48.2% NZ_LQWB01000001.1 Escherichia coli strain GN03624 GCID_EC...
48.0% NZ_CCQJ01000001.1 Escherichia coli strain E. coli, whole ...
47.3% NZ_JHMG01000001.1 Escherichia coli O121:H19 str. 2010EL10...
47.2% NZ_JHGJ01000001.1 Escherichia coli O45:H2 str. 2009C-4780...
46.5% NZ_JHHE01000001.1 Escherichia coli O103:H2 str. 2009C-327...
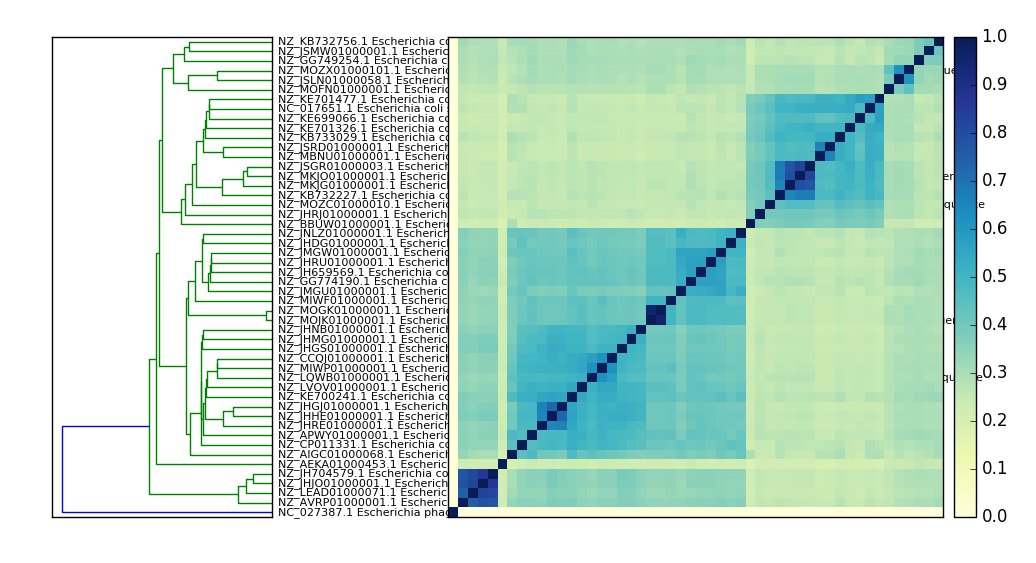
Compare many signatures and build a tree.¶
Compare all the things:
sourmash compare ecoli_many_sigs/* -o ecoli_cmp
Optionally, parallelize to 8 threads using -p 8:
sourmash compare -p 8 ecoli_many_sigs/* -o ecoli_cmp
and then plot:
sourmash plot --pdf --labels ecoli_cmp
which will produce files named ecoli_cmp.matrix.pdf and
ecoli_cmp.dendro.pdf.
Here’s a PNG version:
What’s in my metagenome?¶
Download a database containing all of the GenBank microbial genomes:
curl -L -o genbank-k31.lca.json.gz https://osf.io/4f8n3/download
Next, run the ‘gather’ command to see what’s in your ecoli genome –
sourmash gather ecoli-genome.sig genbank-k31.lca.json.gz
and you should get:
loaded query: /home/diblions/data/ecoliMG165... (k=31, DNA)
loading from genbank-k31.lca.json.gz...
loaded 1 databases.
overlap p_query p_match
--------- ------- -------
4.9 Mbp 100.0% 100.0% LRDF01000001.1 Escherichia coli strai...
found 1 matches total;
the recovered matches hit 100.0% of the query
In this case, the output is kind of boring because this is a single genome. But! You can use this on metagenomes (assembled and unassembled) as well; you’ve just got to make the signature files.
To see this in action, here is gather running on a signature generated from some sequences that assemble (but don’t align to known genomes) from the Shakya et al. 2013 mock metagenome paper.
wget https://github.com/sourmash-bio/sourmash/raw/latest/doc/_static/shakya-unaligned-contigs.sig
sourmash gather -k 31 shakya-unaligned-contigs.sig genbank-k31.lca.json.gz
This should yield:
loaded query: mqc500.QC.AMBIGUOUS.99.unalign... (k=31, DNA)
loaded 1 databases.
overlap p_query p_match
--------- ------- -------
1.4 Mbp 11.0% 58.0% JANA01000001.1 Fusobacterium sp. OBRC...
1.0 Mbp 7.7% 25.9% CP001957.1 Haloferax volcanii DS2 pla...
0.9 Mbp 7.4% 11.8% BA000019.2 Nostoc sp. PCC 7120 DNA, c...
0.7 Mbp 5.9% 23.0% FOVK01000036.1 Proteiniclasticum rumi...
0.7 Mbp 5.3% 17.6% AE017285.1 Desulfovibrio vulgaris sub...
0.6 Mbp 4.9% 11.1% CP001252.1 Shewanella baltica OS223, ...
0.6 Mbp 4.8% 27.3% AP008226.1 Thermus thermophilus HB8 g...
0.6 Mbp 4.4% 11.2% CP000031.2 Ruegeria pomeroyi DSS-3, c...
480.0 kbp 3.8% 7.6% CP000875.1 Herpetosiphon aurantiacus ...
410.0 kbp 3.3% 10.5% CH959317.1 Sulfitobacter sp. NAS-14.1...
1.4 Mbp 2.2% 11.8% LN831027.1 Fusobacterium nucleatum su...
0.5 Mbp 2.1% 5.3% CP000753.1 Shewanella baltica OS185, ...
420.0 kbp 1.9% 7.7% FNDZ01000023.1 Proteiniclasticum rumi...
150.0 kbp 1.2% 4.6% AE000513.1 Deinococcus radiodurans R1...
150.0 kbp 1.2% 8.2% CP000969.1 Thermotoga sp. RQ2, comple...
290.0 kbp 1.1% 4.1% CH959311.1 Sulfitobacter sp. EE-36 sc...
1.2 Mbp 1.0% 5.0% CP013328.1 Fusobacterium nucleatum su...
110.0 kbp 0.9% 3.7% FRDZ01000215.1 Enterococcus faecalis ...
0.6 Mbp 0.8% 2.8% CP000527.1 Desulfovibrio vulgaris DP4...
70.0 kbp 0.6% 1.2% CP000850.1 Salinispora arenicola CNS-...
340.0 kbp 0.6% 3.3% KQ235732.1 Fusobacterium nucleatum su...
60.0 kbp 0.5% 0.7% CP000270.1 Burkholderia xenovorans LB...
50.0 kbp 0.4% 2.6% CP001080.1 Sulfurihydrogenibium sp. Y...
50.0 kbp 0.4% 3.2% L77117.1 Methanocaldococcus jannaschi...
found less than 40.0 kbp in common. => exiting
found 24 matches total;
the recovered matches hit 73.1% of the query
If you use the -o flag, gather will write out a csv that contains additional information. The column headers and their meanings are:
intersect_bp: the approximate number of base pairs in common between the query and the matchf_orig_query: fraction of original query; the fraction of the original query that is contained within the matchf_match: fraction of match; the fraction of the match that is contained within the queryf_unique_to_query: fraction unique to query; the fraction of the query that uniquely overlaps with the matchf_unique_weighted: fraction unique to query weighted by abundance; fraction unique to query, weighted by abundance in the query
It is straightforward to build your own databases for use with search
and gather; see sourmash index, above, the LCA tutorial, or
our notebook on working with private collections of signatures.
