Building plots from sourmash compare output¶
Running this notebook.¶
You can run this notebook interactively via mybinder; click on this button:
A rendered version of this notebook is available at sourmash.readthedocs.io under “Tutorials and notebooks”.
You can also get this notebook from the doc/ subdirectory of the sourmash github repository. See binder/environment.yaml for installation dependencies.
What is this?¶
This is a Jupyter Notebook using Python 3. If you are running this via binder, you can use Shift-ENTER to run cells, and double click on code cells to edit them.
Contact: C. Titus Brown, ctbrown@ucdavis.edu. Please file issues on GitHub if you have any questions or comments!
Running sourmash compare and generating figures in Python¶
First, we need to generate a similarity matrix with compare. (If you want to generate this programmatically, it’s just a numpy matrix.)
[1]:
!sourmash compare ../tests/test-data/demo/*.sig -o compare-demo
== This is sourmash version 4.8.5.dev0. ==
== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==
loaded 7 signatures total.
0-SRR2060939_1.fa... [1. 0.356 0.078 0.086 0. 0. 0. ]
1-SRR2060939_2.fa... [0.356 1. 0.072 0.078 0. 0. 0. ]
2-SRR2241509_1.fa... [0.078 0.072 1. 0.074 0. 0. 0. ]
3-SRR2255622_1.fa... [0.086 0.078 0.074 1. 0. 0. 0. ]
4-SRR453566_1.fas... [0. 0. 0. 0. 1. 0.382 0.364]
5-SRR453569_1.fas... [0. 0. 0. 0. 0.382 1. 0.386]
6-SRR453570_1.fas... [0. 0. 0. 0. 0.364 0.386 1. ]
min similarity in matrix: 0.000
saving labels to: compare-demo.labels.txt
saving comparison matrix to: compare-demo
[2]:
%pylab inline
# import the `fig` module from sourmash:
from sourmash import fig
%pylab is deprecated, use %matplotlib inline and import the required libraries.
Populating the interactive namespace from numpy and matplotlib
The sourmash.fig module contains code to load the similarity matrix and associated labels:
[3]:
matrix, labels = fig.load_matrix_and_labels('compare-demo')
Here, matrix is a numpy matrix and labels is a list of labels (by default, filenames).
[4]:
print('matrix:\n', matrix)
print('labels:', labels)
matrix:
[[1. 0.356 0.078 0.086 0. 0. 0. ]
[0.356 1. 0.072 0.078 0. 0. 0. ]
[0.078 0.072 1. 0.074 0. 0. 0. ]
[0.086 0.078 0.074 1. 0. 0. 0. ]
[0. 0. 0. 0. 1. 0.382 0.364]
[0. 0. 0. 0. 0.382 1. 0.386]
[0. 0. 0. 0. 0.364 0.386 1. ]]
labels: ['SRR2060939_1.fastq.gz', 'SRR2060939_2.fastq.gz', 'SRR2241509_1.fastq.gz', 'SRR2255622_1.fastq.gz', 'SRR453566_1.fastq.gz', 'SRR453569_1.fastq.gz', 'SRR453570_1.fastq.gz']
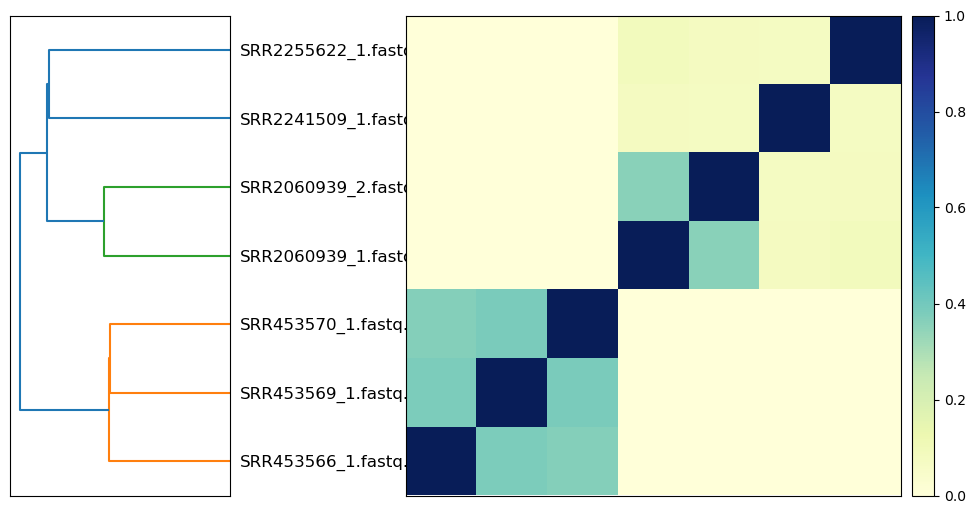
The plot_composite_matrix function returns a generated plot, along with the labels and matrix as re-ordered by the clustering:
[5]:
f, reordered_labels, reordered_matrix = fig.plot_composite_matrix(matrix, labels)
[6]:
print('reordered matrix:\n', reordered_matrix)
print('reordered labels:', reordered_labels)
reordered matrix:
[[1. 0.382 0.364 0. 0. 0. 0. ]
[0.382 1. 0.386 0. 0. 0. 0. ]
[0.364 0.386 1. 0. 0. 0. 0. ]
[0. 0. 0. 1. 0.356 0.078 0.086]
[0. 0. 0. 0.356 1. 0.072 0.078]
[0. 0. 0. 0.078 0.072 1. 0.074]
[0. 0. 0. 0.086 0.078 0.074 1. ]]
reordered labels: ['SRR453566_1.fastq.gz', 'SRR453569_1.fastq.gz', 'SRR453570_1.fastq.gz', 'SRR2060939_1.fastq.gz', 'SRR2060939_2.fastq.gz', 'SRR2241509_1.fastq.gz', 'SRR2255622_1.fastq.gz']
Customizing plots¶
If you want to customize the plots, please see the code for plot_composite_matrix in sourmash/fig.py, which is reproduced below; you can modify the code in place to (for example) use custom dendrogram colors.
[7]:
import scipy.cluster.hierarchy as sch
def plot_composite_matrix(D, labeltext, show_labels=True,
vmax=1.0, vmin=0.0, force=False):
"""Build a composite plot showing dendrogram + distance matrix/heatmap.
Returns a matplotlib figure.
If show_labels is True, display labels. Otherwise, no labels are
shown on the plot.
"""
if D.max() > 1.0 or D.min() < 0.0:
error('This matrix doesn\'t look like a distance matrix - min value {}, max value {}', D.min(), D.max())
if not force:
raise ValueError("not a distance matrix")
else:
notify('force is set; scaling to [0, 1]')
D -= D.min()
D /= D.max()
if show_labels:
show_indices = True
fig = pylab.figure(figsize=(11, 8))
ax1 = fig.add_axes([0.09, 0.1, 0.2, 0.6])
# plot dendrogram
Y = sch.linkage(D, method='single') # centroid
Z1 = sch.dendrogram(Y, orientation='left', labels=labeltext,
no_labels=not show_labels, get_leaves=True)
ax1.set_xticks([])
xstart = 0.45
width = 0.45
if not show_labels:
xstart = 0.315
scale_xstart = xstart + width + 0.01
# re-order labels along rows, top to bottom
idx1 = Z1['leaves']
reordered_labels = [ labeltext[i] for i in idx1 ]
# reorder D by the clustering in the dendrogram
D = D[idx1, :]
D = D[:, idx1]
# show matrix
axmatrix = fig.add_axes([xstart, 0.1, width, 0.6])
im = axmatrix.matshow(D, aspect='auto', origin='lower',
cmap=pylab.cm.YlGnBu, vmin=vmin, vmax=vmax)
axmatrix.set_xticks([])
axmatrix.set_yticks([])
# Plot colorbar.
axcolor = fig.add_axes([scale_xstart, 0.1, 0.02, 0.6])
pylab.colorbar(im, cax=axcolor)
return fig, reordered_labels, D
[8]:
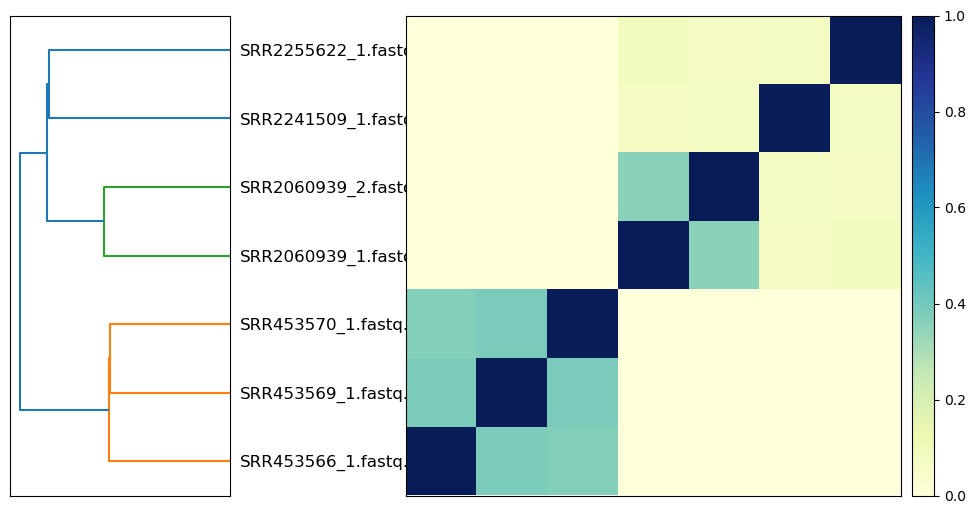
_ = plot_composite_matrix(matrix, labels)
[ ]:
